 |
The geography of novel and atypical research
Q Ke, Tianxing Pan, J Mao
Research Policy 55, 105345 (2026)
arXiv |
abstract |
BibTeX |
data & code
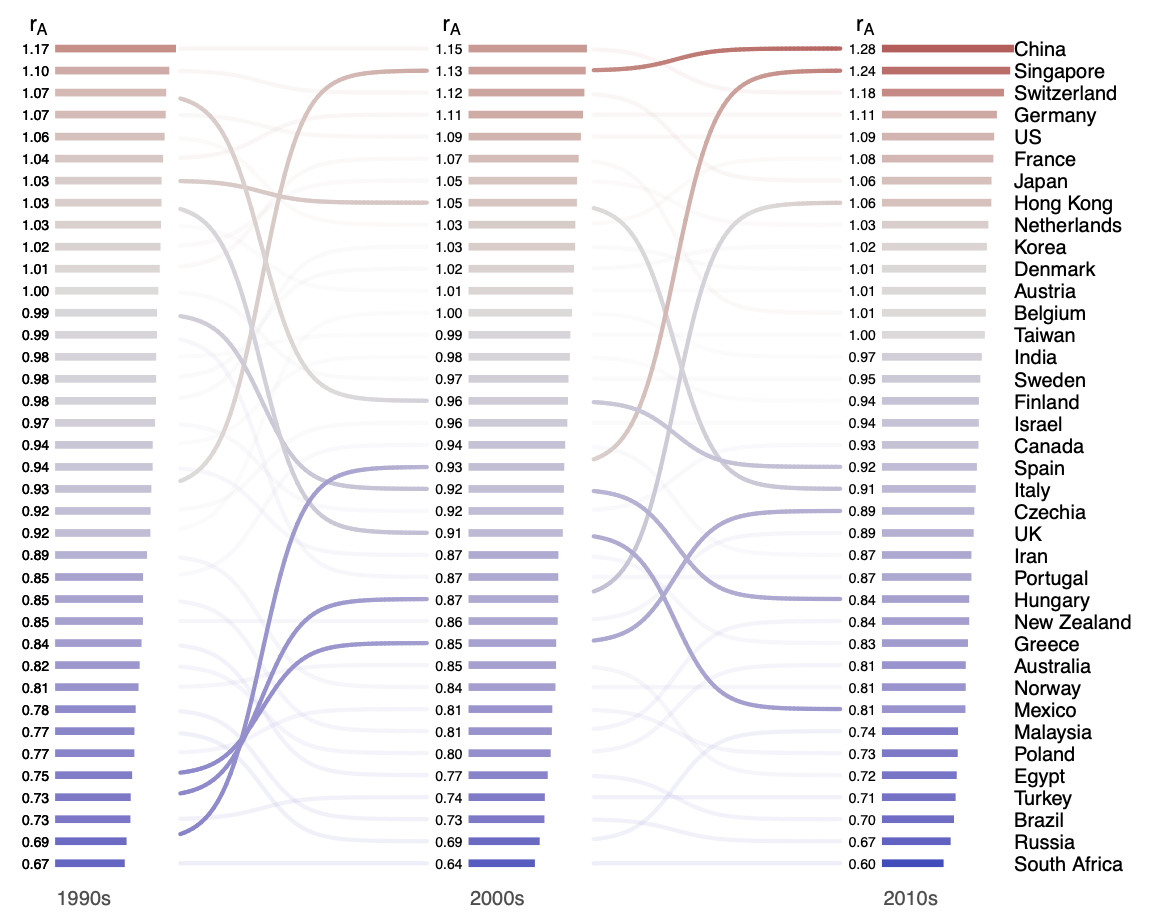
The production of knowledge has become increasingly a global endeavor. Yet, location related factors, such as local working environment and national policy designs, may continue to affect what kind of science is being pursued. Here we examine the geography of the production of creative science by country, through the lens of novelty and atypicality proposed in Uzzi et al. (2013). We quantify a country’s representativeness in novel and atypical science, finding persistent differences in propensity to generate creative works, even among developed countries that are large producers in science. We further cluster countries based on how their tendency to publish novel science changes over time, identifying one group of emerging countries. Our analyses point out the recent emergence of China not only as a large producer in science but also as a leader that disproportionately produces more novel and atypical research. Discipline specific analysis indicates that China’s over-production of atypical science is limited to a few disciplines, especially its most prolific ones like materials science and chemistry.
@article{ke2025geography,
title={The geography of novel and atypical research},
author={Ke, Qing and Pan, Tianxing and Mao, Jin},
journal={Research Policy},
volume={55},
pages={105345},
year={2026},
doi={10.1016/j.respol.2025.105345}
}
|
 |
Mapping the changing structure of science through diachronic periodical embeddings
Zhuoqi Lyu, Q Ke
Chaos, Solitons & Fractals 201, 117295 (2025)
arXiv |
abstract |
BibTeX |
code
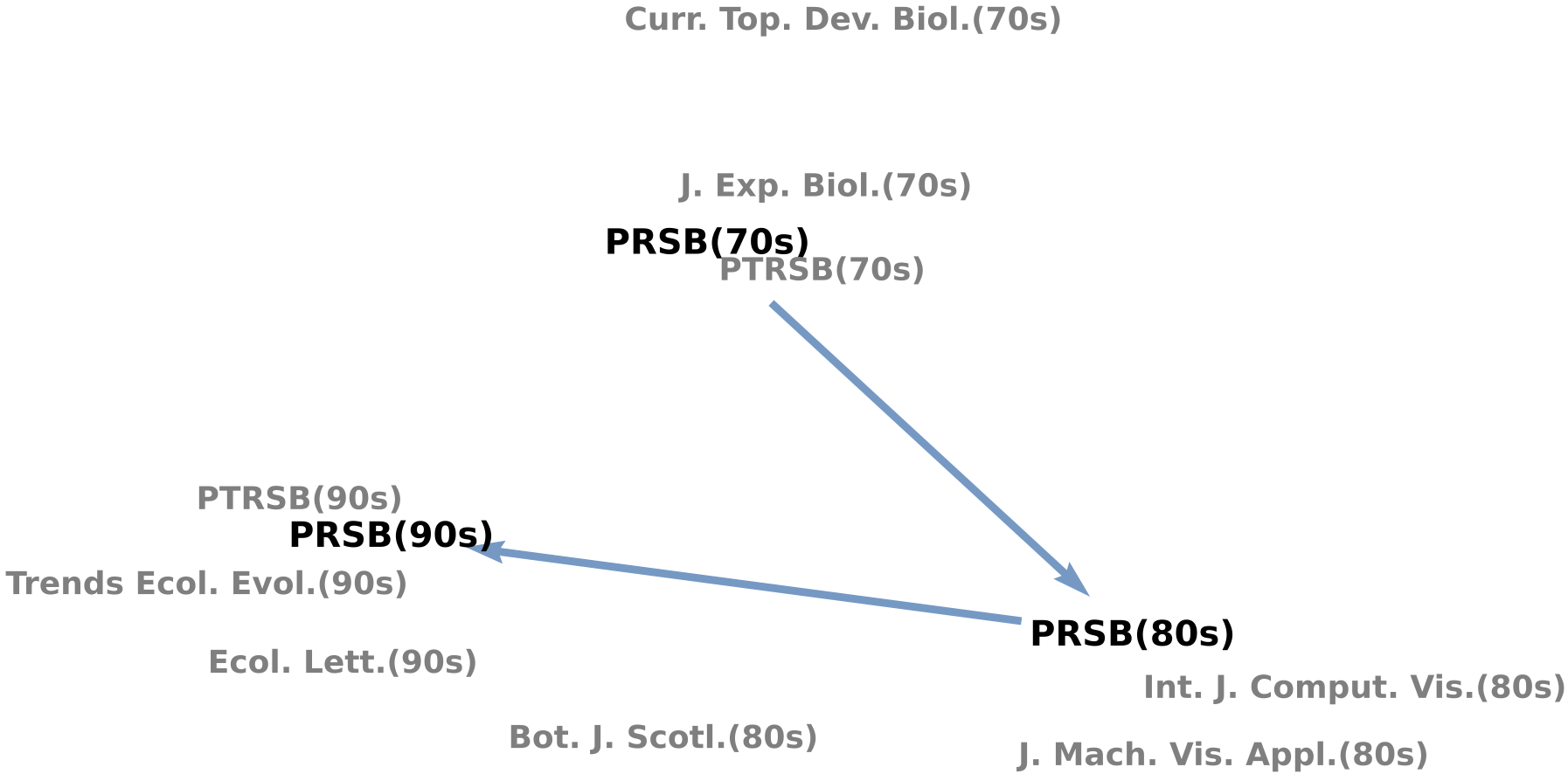
Charting the changing landscape of scientific research is a fundamental challenge in the science of science. We develop diachronic embeddings of scholarly periodicals to quantify “semantic changes” of periodicals across decades, allowing us to track the evolution of research topics and identify rapidly developing fields. By mapping periodicals within a physical-life-health triangle, we reveal an evolving interdisciplinary science landscape, finding an overall trend toward specialization for most periodicals but increasing interdisciplinarity for bioscience periodicals. Analyzing a periodical’s trajectory within this triangle over time allows us to visualize how its research focus shifts. Furthermore, by monitoring the formation of local clusters of periodicals, we can identify emerging research topics such as AIDS research and nanotechnology in the 1980s. Our work offers novel quantification in the science of science and provides a quantitative lens to examine the evolution of science, which may facilitate future investigations into the emergence and development of research fields.
@article{lyu2025mapping,
title={Mapping the changing structure of science through diachronic periodical embeddings},
author={Lyu, Zhuoqi and Ke, Qing},
journal={Chaos, Solitons \& Fractals},
volume={201},
pages={117295},
year={2025},
doi={10.1016/j.chaos.2025.117295}
}
|
![]() |
Help me screen: Analyzing and predicting the success of start-ups in dynamic venture capital networks
S Lyu, X Li, S Hong, Q Ke, J Gu, K Zhang, H Zhang
ACM Transactions on Intelligent Systems and Technology 16, 129 (2025)
arXiv |
abstract |
BibTeX
Most start-ups fail, and early-stage ventures face even lower survival rates. Identifying high-potential start-ups remains a critical challenge for venture capital (VC) investors and policymakers. While predictive models exist, the evolving relationships between VC investors, start-ups, and management teams in dynamic networks are underexplored. We propose a method to predict whether a start-up will succeed within five years of its first funding round. Using a 40-year global VC dataset, we model the VC ecosystem as a dynamic bipartite network linking start-ups to individuals (investors/managers). Our approach incrementally updates graph embeddings through unsupervised self-attention to incorporate new nodes, edges, and their neighbors. Node embeddings are further fine-tuned via link prediction and classification tasks, while temporal dependencies are captured to form sequential representations. The model identifies early-stage start-ups with twice the success likelihood of those chosen by professional investors. Key factors including networking and education align with VC literature. Additionally, we provide model complexity analysis and open-source our implementation to support practical applications and future research.
@article{lyu2025help,
title={Help Me Screen: Analyzing and Predicting the Success of Start-ups in Dynamic Venture Capital Networks},
author={Lyu, Shiwei and Li, Xiaofeng and Hong, Suting and Ke, Qing and Gu, Jinjie and Zhang, Kunpeng and Zhang, Haipeng},
journal={ACM Transactions on Intelligent Systems and Technology},
volume={16},
number={6},
articleno={129},
year={2025},
doi={10.1145/3763001}
}
|
![]() |
Decoding the writing styles of disciplines: A large-scale quantitative analysis
S Dong, J Mao, Q Ke, L Pei
Information Processing & Management 61, 103718 (2024)
abstract |
BibTeX
Disciplinary writing style stems from the practice of science, reflecting the scientific culture. This study aims to explore the differences and evolution of scientific writing styles from the perspective of disciplines. A large-scale quantitative analysis was conducted over 14 million abstracts from the Microsoft Academic Graph (MAG) database across eight soft and hard disciplines. Represented by a comprehensive set of 14 symbolic, lexical, syntactic, structural, and readability features, the evolution of disciplinary writing styles was analyzed over 30 years. Interpretable machine learning methods were performed to test the discernibility of writing styles across disciplines and disclose their linguistic differences. Our findings reveal the linguistic features of soft disciplines (Art, Philosophy, and Sociology) and Mathematics generally keep stabilized, and a general trend of increasing linguistic complexity was observed for Biology, Chemistry, Computer Science, and Psychology. The good performance of the pairwise writing style classifiers indicates a well discriminability of the writing styles between disciplines. A correlation between the performance of classifiers and the distance between disciplines was identified. The feature contribution analysis using SHapley Additive exPlanations (SHAP) and Kendall's Tau rank correlation revealed the detailed commonalities and disparities in disciplines’ linguistic features. This study provides profound insights into the understanding of scientific writing and norms, which further helps develop useful tools for academic text analysis, foster interdisciplinary communication, and assist educators to construct discipline-specific writing guidance.
@article{dong2024decoding,
title={Decoding the writing styles of disciplines: A large-scale quantitative analysis},
author={Dong, Shuyi and Mao, Jin and Ke, Qing and Pei, Lei},
journal={Information Processing \& Management},
volume={61},
number={4},
pages={103718},
year={2024},
doi={10.1016/j.ipm.2024.103718}
}
|
 |
A network-based normalized impact measure reveals successful periods of scientific discovery across disciplines
Q Ke, AJ Gates, AL Barabási
PNAS 120, e2309378120 (2023)
abstract |
BibTeX |
cover
The impact of a scientific publication is often measured by the number of citations it receives from the scientific community. However, citation count is susceptible to well-documented variations in citation practices across time and discipline, limiting our ability to compare different scientific achievements. Previous efforts to account for citation variations often rely on a priori discipline labels of papers, assuming that all papers in a discipline are identical in their subject matter. Here, we propose a network-based methodology to quantify the impact of an article by comparing it with locally comparable research, thereby eliminating the discipline label requirement. We show that the developed measure is not susceptible to discipline bias and follows a universal distribution for all articles published in different years, offering an unbiased indicator for impact across time and discipline. We then use the indicator to identify science-wide high impact research in the past half century and quantify its temporal production dynamics across disciplines, helping us identifying breakthroughs from diverse, smaller disciplines, such as geosciences, radiology, and optics, as opposed to citation-rich biomedical sciences. Our work provides insights into the evolution of science and paves a way for fair comparisons of the impact of diverse contributions across many fields.
@article{ke2023network,
title={A network-based normalized impact measure reveals successful periods of scientific discovery across discipline},
author={Ke, Qing and Gates, Alexander J and Barab{\'a}si, Albert-L{\'a}szl{\'o}},
journal={Proceedings of the National Academy of Sciences},
volume={120},
pages={e2309378120},
year={2023},
doi={10.1073/pnas.2309378120}
}
AJ Gates, Q Ke, & AL Barabási (2024). Reply to Vaccario et al.: The role of baselines in fair and unbiased citation metric evaluation. PNAS, 121, e2410675121. -- The first law of bibliometrics: number of papers is growing exponentially every year.
|
 |
Interdisciplinary research and technological impact: Evidence from biomedicine
Q Ke
Scientometrics 128, 2035–2077 (2023)
arXiv |
abstract |
BibTeX
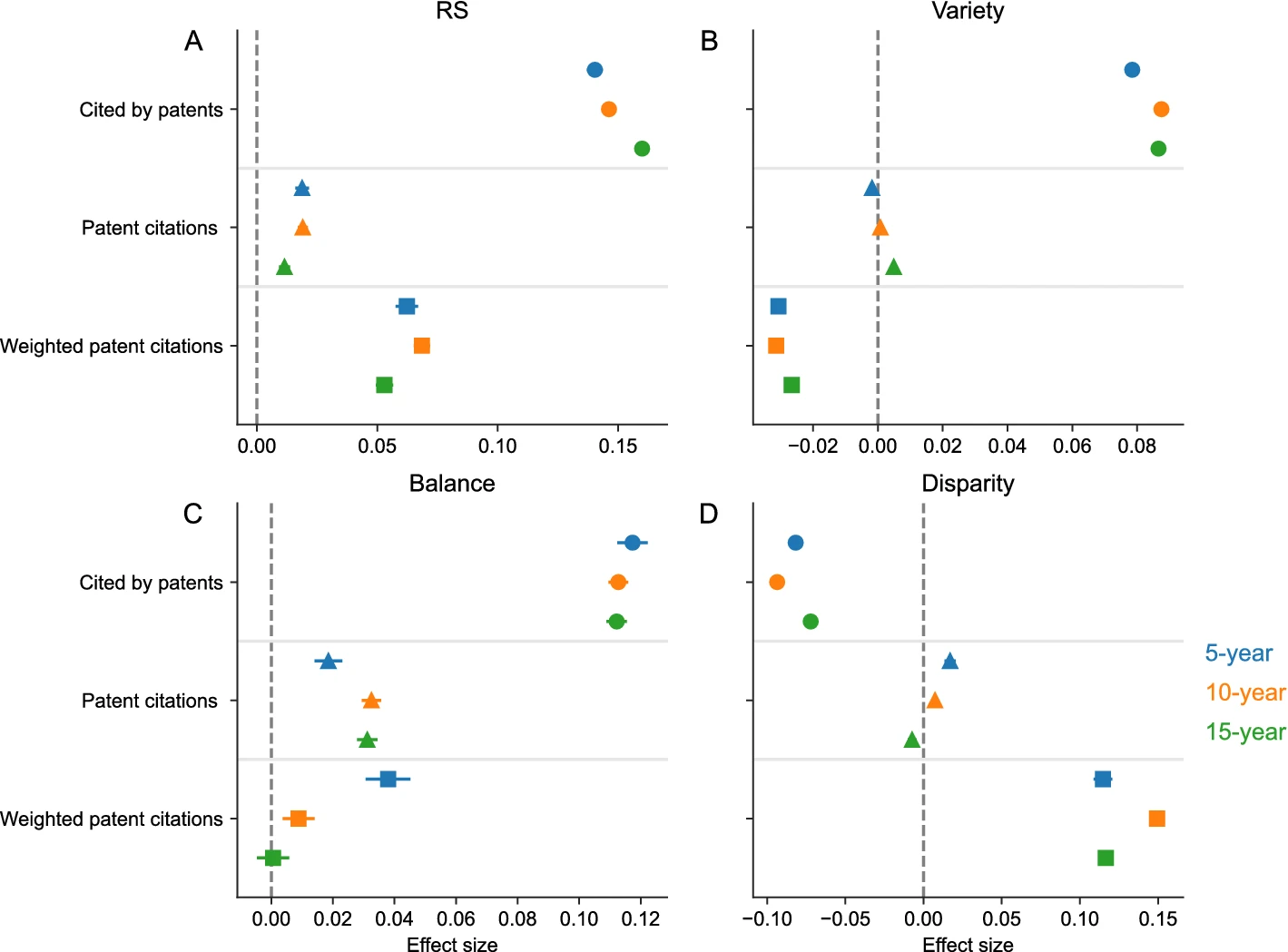
Interdisciplinary research (IDR) has been considered as an important source for scientific breakthroughs and as a solution to today’s complex societal challenges. While ample empirical evidence has suggested its benefits within the academia such as better creativity and higher scientific impact and visibility, its societal benefits—a key argument originally used for promoting IDR—remain relatively unexplored. Here, we study one aspect of societal benefits, that is contributing to the development of patented technologies, and examine how IDR papers are referenced as “prior art” by patents over time. We draw on a large sample of biomedical papers published in 23 years and measure the degree of interdisciplinarity of a paper using three popular indicators, namely variety, balance, and disparity. We find that papers that cites more fields (variety) and whose distributions over those cited fields are more even (balance) are more likely to receive patent citations, but both effects can be offset if papers draw upon more distant fields (disparity). These associations are consistent across different citation-window lengths. We further find that conditional on receiving patent citations, the intensity of their technological impact, as measured as both raw and quality-adjusted number of citing patents, increases with balance and disparity. Our work may have policy implications for interdisciplinary research and scientific and technological impact.
@article{ke2023interdisciplinary,
title={Interdisciplinary research and technological impact: evidence from biomedicine},
author={Ke, Qing},
journal={Scientometrics},
volume={128},
pages={2035--2077},
year={2023},
doi={10.1007/s11192-023-04662-0}
}
|
 |
A dataset of mentorship in bioscience with semantic and demographic estimations
Q Ke, L Liang, Y Ding, SV David, DE Acuna
Scientific Data 9, 467 (2022)
arXiv |
abstract |
BibTeX |
Data |
Code
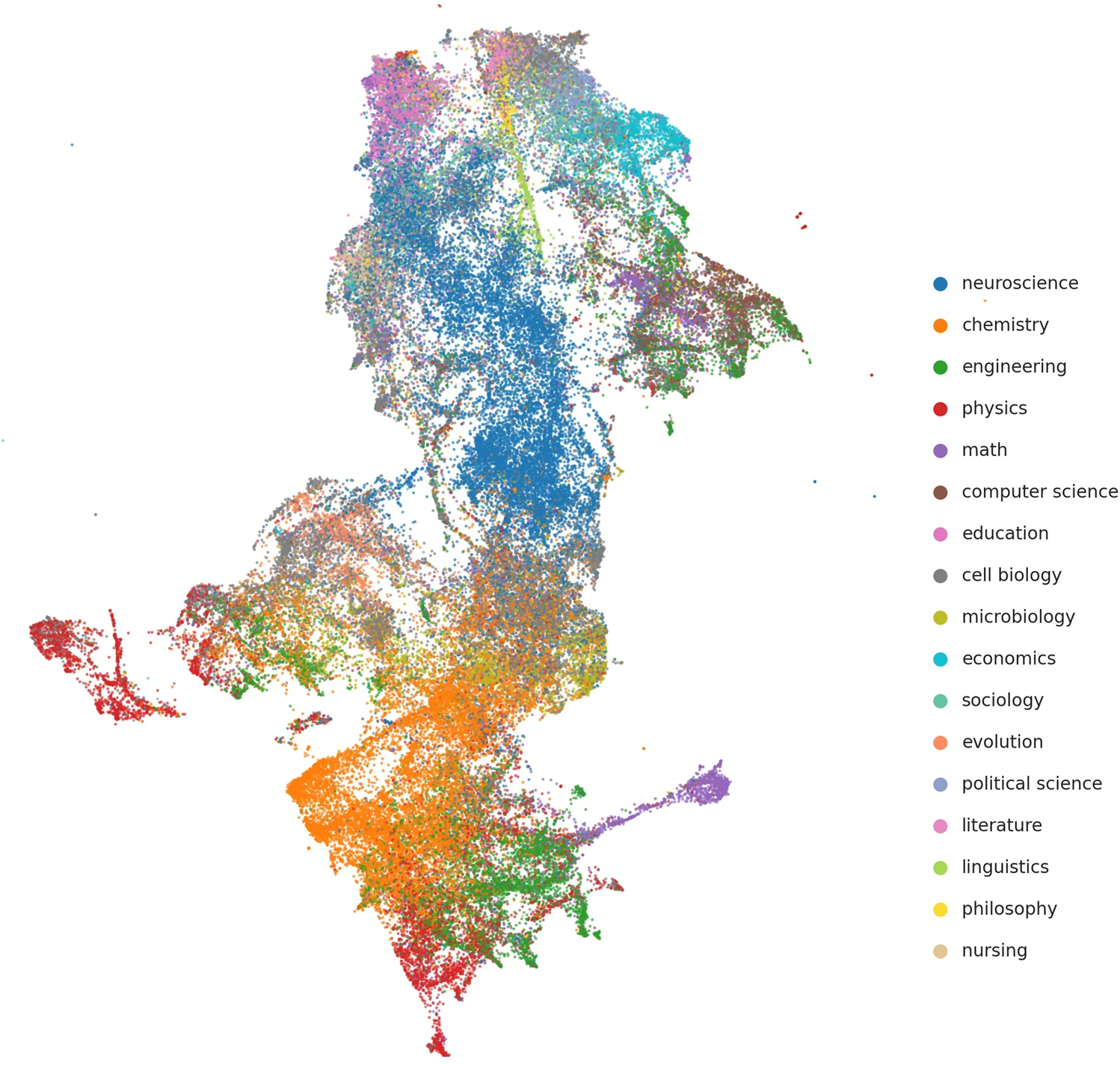
Mentorship in science is crucial for topic choice, career decisions, and the success of mentees and mentors. Typically, researchers who study mentorship use article co-authorship and doctoral dissertation datasets. However, available datasets of this type focus on narrow selections of fields and miss out on early career and non-publication-related interactions. Here, we describe Mentorship, a crowdsourced dataset of 743176 mentorship relationships among 738989 scientists primarily in biosciences that avoids these shortcomings. Our dataset enriches the Academic Family Tree project by adding publication data from the Microsoft Academic Graph and “semantic” representations of research using deep learning content analysis. Because gender and race have become critical dimensions when analyzing mentorship and disparities in science, we also provide estimations of these factors. We perform extensive validations of the profile–publication matching, semantic content, and demographic inferences, which mostly cover neuroscience and biomedical sciences. We anticipate this dataset will spur the study of mentorship in science and deepen our understanding of its role in scientists’ career outcomes.
@article{ke2022dataset,
title={A dataset of mentorship in bioscience with semantic and demographic estimations},
author={Ke, Qing and Liang, Lizhen and Ding, Ying and David, Stephen V and Acuna, Daniel E},
journal={Scientific data},
volume={9},
pages={467},
year={2022},
doi={10.1038/s41597-022-01578-x}
}
|
 |
Neural embeddings of scholarly periodicals reveal complex disciplinary organizations
H Peng, Q Ke, C Budak, DM Romero, YY Ahn
Science Advances 7, eabb9004 (2021)
arXiv |
abstract |
BibTeX |
Data |
Code
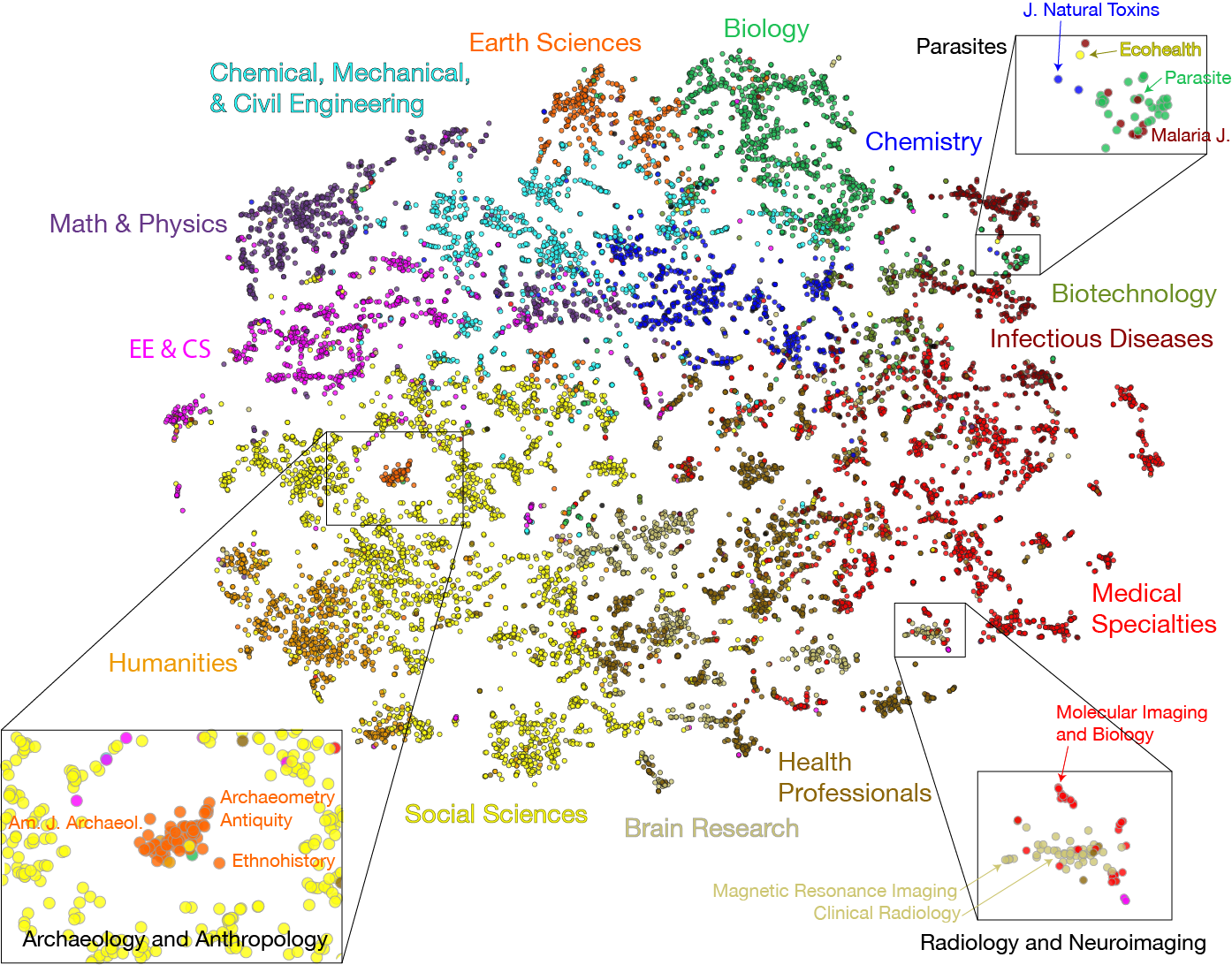
Understanding the structure of knowledge domains is one of the foundational challenges in the science of science. Here, we propose a neural embedding technique that leverages the information contained in the citation network to obtain continuous vector representations of scientific periodicals. We demonstrate that our periodical embeddings encode nuanced relationships between periodicals and the complex disciplinary and interdisciplinary structure of science, allowing us to make cross-disciplinary analogies between periodicals. Furthermore, we show that the embeddings capture meaningful “axes” that encompass knowledge domains, such as an axis from “soft” to “hard” sciences or from “social” to “biological” sciences, which allow us to quantitatively ground periodicals on a given dimension. By offering novel quantification in the science of science, our framework may, in turn, facilitate the study of how knowledge is created and organized.
@article{peng2021neural,
author = {Peng, Hao and Ke, Qing and Budak, Ceren and Romero, Daniel M. and Ahn, Yong-Yeol},
title = {Neural embeddings of scholarly periodicals reveal complex disciplinary organizations},
journal = {Science Advances},
volume = {7},
number = {17},
pages = {eabb9004},
year = {2021},
doi = {10.1126/sciadv.abb9004}
}
|
 |
An analysis of the evolution of science-technology linkage in biomedicine
Q Ke
Journal of Informetrics 14, 101074 (2020)
arXiv |
abstract |
BibTeX
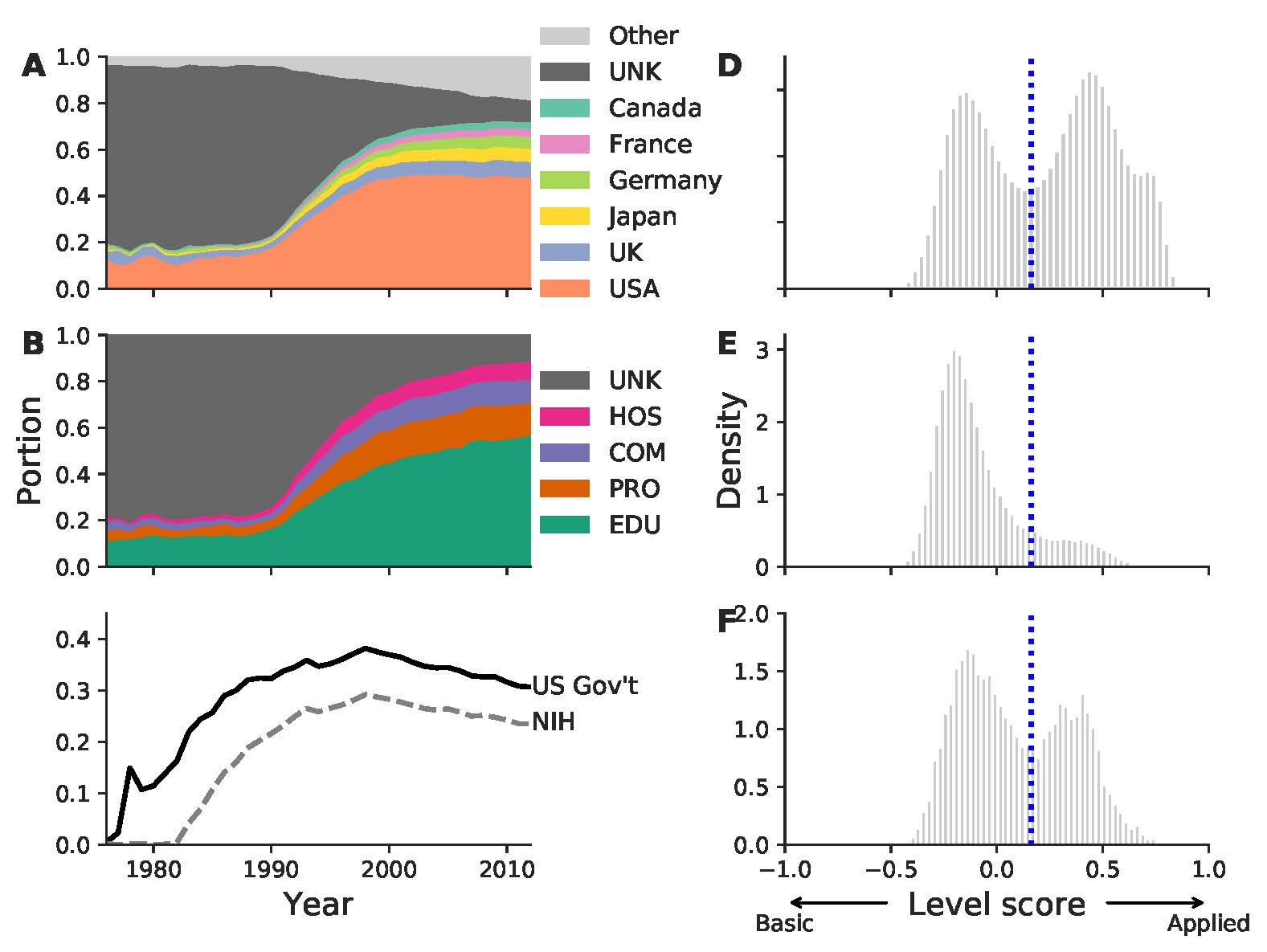
Demonstrating the practical value of public research has been an important subject in science policy. Here we present a detailed study on the evolution of the citation linkage between life science related patents and biomedical research over a 37-year period. Our analysis relies on a newly-created dataset that systematically links millions of non-patent references to biomedical papers. We find a large disparity in the volume of citations to science among technology sectors, with biotechnology and drug patents dominating it. The linkage has been growing exponentially over a long period of time, doubling every 2.9 years. The U.S. has been the largest producer of cited science for years, receiving nearly half of the citations. More than half of citations goes to universities. We use a new paper-level indicator to quantify to what extent a paper is basic research or clinical medicine. We find that the cited papers are likely to be basic research, yet a significant portion of papers cited in patents that are related to FDA-approved drugs are clinical research. The U.S. National Institute of Health continues to be an important funder of cited science. For the majority of companies, more than half of citations in their patents are authored by public research. Taken together, these results indicate a continuous linkage of public science to private sector inventions.
@article{ke2020analysis,
author = {Qing Ke},
title = {An analysis of the evolution of science-technology linkage in biomedicine},
journal = {Journal of Informetrics},
volume = {14},
pages = {101074},
year = {2020},
doi = {10.1016/j.joi.2020.101074}
}
|
 |
Technological impact of biomedical research: The role of basicness and novelty
Q Ke
Research Policy 49, 104071 (2020)
arXiv |
abstract |
BibTeX
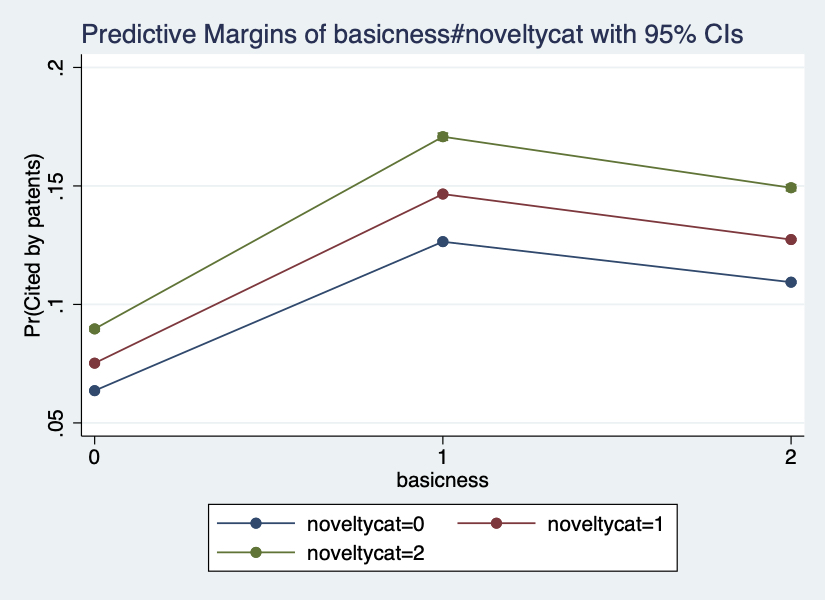
An ongoing interest in innovation studies is to understand how knowledge generated from scientific research can be used in the development of technologies. While previous inquiries have devoted to studying the scientific capacity of technologies and institutional factors facilitating technology transfer, little is known about the intrinsic characteristics of scientific publications that gain direct technological impact. Here we focus on two features, namely basicness and novelty. Using a corpus of 3.8 million papers published between 1980 and 1999, we find that basic science papers and novel papers are substantially more likely to achieve direct technological impact. Further analysis that limits to papers with technological impact reveals that basic science and novel science have more patent citations, experience shorter time lag, and have impact in broader technological fields.
@article{ke2020technological,
author = {Qing Ke},
title = {Technological impact of biomedical research: The role of basicness and novelty},
journal = {Research Policy},
volume = {49},
pages = {104071},
year = {2020},
doi = {10.1016/j.respol.2020.104071}
}
|
 |
The citation disadvantage of clinical research
Q Ke
Journal of Informetrics 14, 100998 (2020)
arXiv |
abstract |
BibTeX
Biomedical research encompasses diverse types of activities, from basic science (“bench”) to clinical medicine (“bedside”) to bench-to-bedside translational research. It, however, remains unclear whether different types of research receive citations at varying rates. Here we aim to answer this question by using a newly proposed paper-level indicator that quantifies the extent to which a paper is basic science or clinical medicine. Applying this measure to 5 million biomedical papers, we find a systematic citation disadvantage of clinical oriented papers; they tend to garner far fewer citations and are less likely to be hit works than papers oriented towards basic science. At the same time, clinical research has a higher variance in its citation. We also find that the citation difference between basic and clinical research decreases, yet still persists, if longer citation-window is used. Given the increasing adoption of short-term, citation-based bibliometric indicators in funding decisions, the under-cited effect of clinical research may provide disincentives for bio-researchers to venture into the translation of basic scientific discoveries into clinical applications, thus providing explanations of reasons behind the existence of the gap between basic and clinical research that is commented as “valley of death” and the commentary of “extinction” risk of translational researchers. Our work may provide insights to policy-makers on how to evaluate different types of biomedical research.
@article{ke2020citation,
author = {Qing Ke},
title = {The citation disadvantage of clinical research},
journal = {Journal of Informetrics},
volume = {14},
pages = {100998},
year = {2020},
doi = {10.1016/j.joi.2019.100998}
}
|
 |
Nature's reach: narrow work has broad impact
AJ Gates, Q Ke, O Varol, AL Barabási
Nature 575, 32-34 (2019)
BibTeX |
video |
cover |
interactive |
2020 Webby Awards for NetArt
@article{gates2019nature,
author = {Alexander J. Gates and Qing Ke and Onur Varol and Albert-László Barabási},
title = {Nature's reach: narrow work has broad impact},
journal = {Nature},
volume = {575},
pages = {32--34},
year = {2019},
doi = {10.1038/d41586-019-03308-7}
}
|
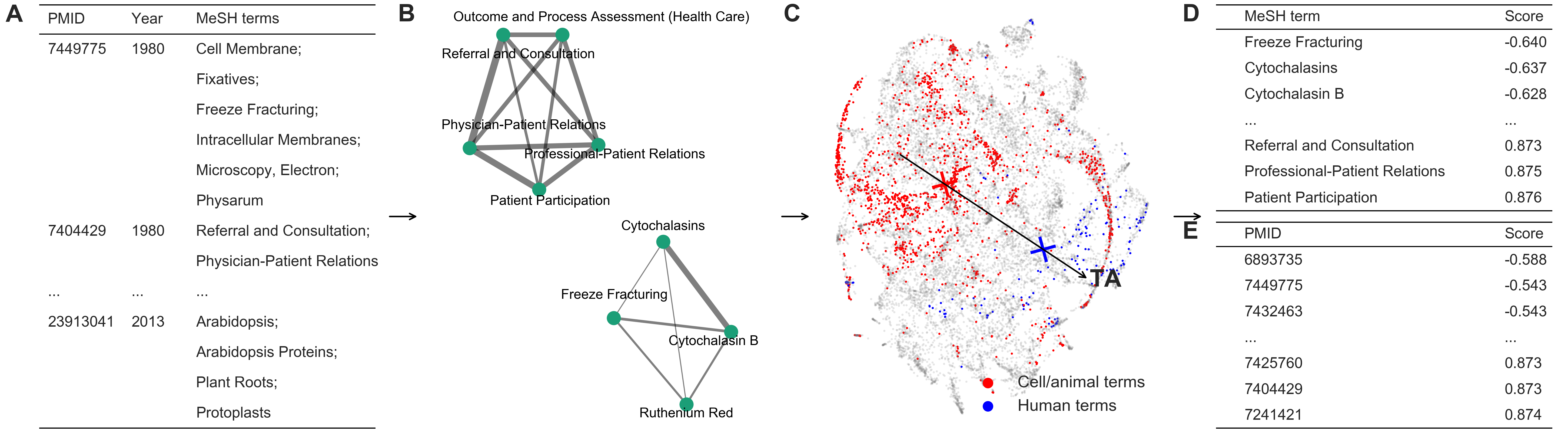 |
Identifying translational science through embeddings of controlled vocabularies
Q Ke
Journal of the American Medical Informatics Association 26, 516-523 (2019)
arXiv |
abstract |
BibTeX |
SI |
Data |
Code
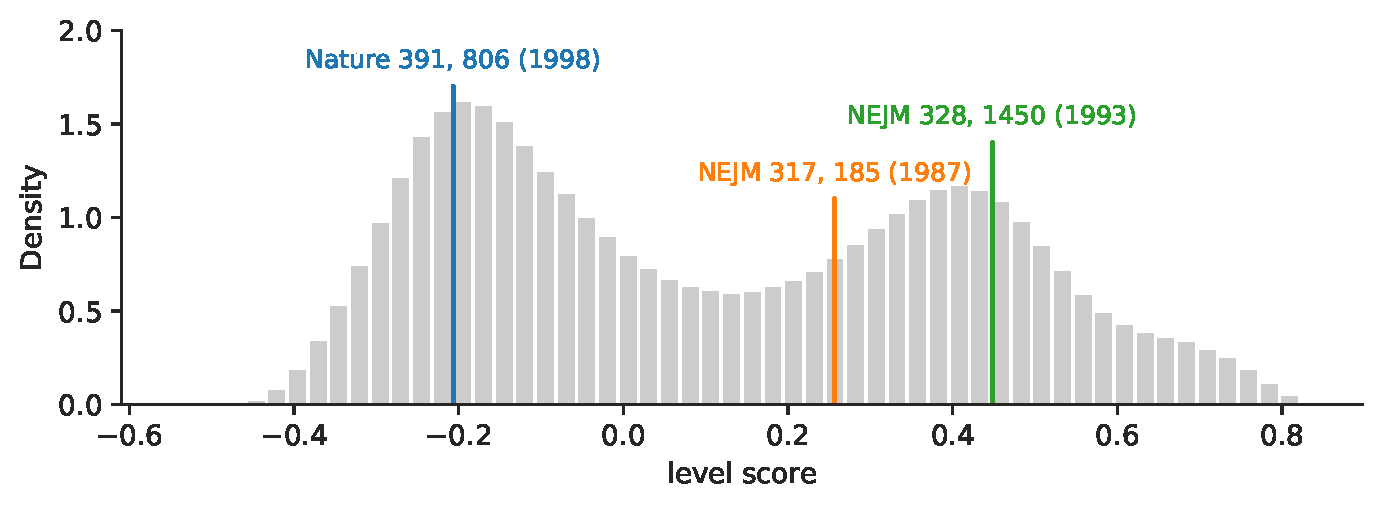
Objective: Translational science aims at "translating" basic scientific discoveries into clinical applications. The identification of translational science has practicality such as evaluating the effectiveness of investments made into large programs like the Clinical and Translational Science Awards. Despite several proposed methods that group publications—the primary unit of research output—into some categories, we still lack a quantitative way to place papers onto the full, continuous spectrum from basic research to clinical medicine. Methods: Here we learn vector-representations of controlled vocabularies assigned to MEDLINE papers to obtain a Translational Axis (TA) that points from basic science to clinical medicine. The projected position of a term on the TA, expressed by a continuous quantity, indicates the term's "appliedness." The position of a paper, determined by the average location over its terms, quantifies the degree of its "appliedness," which we term as "level score." Results: We validate our method by comparing with previous techniques, showing excellent agreement yet uncovering significant variations of scores of papers in previously defined categories. The measure allows us to characterize the standing of journals, disciplines, and the entire biomedical literature along the basic-applied spectrum. Analysis on large-scale citation network reveals two main findings. First, direct citations mainly occurred between papers with similar scores. Second, shortest paths are more likely ended up with a paper closer to the basic end of the spectrum, regardless of where the starting paper is on the spectrum. Conclusions: The proposed method provides a quantitative way to identify translational science.
@article{ke2019identifying,
author = {Qing Ke},
title = {Identifying translational science through embeddings of controlled vocabularies},
journal = {Journal of the American Medical Informatics Association},
volume = {26},
number = {6},
pages = {516--523},
year = {2019},
doi = {10.1093/jamia/ocy177}
}
|
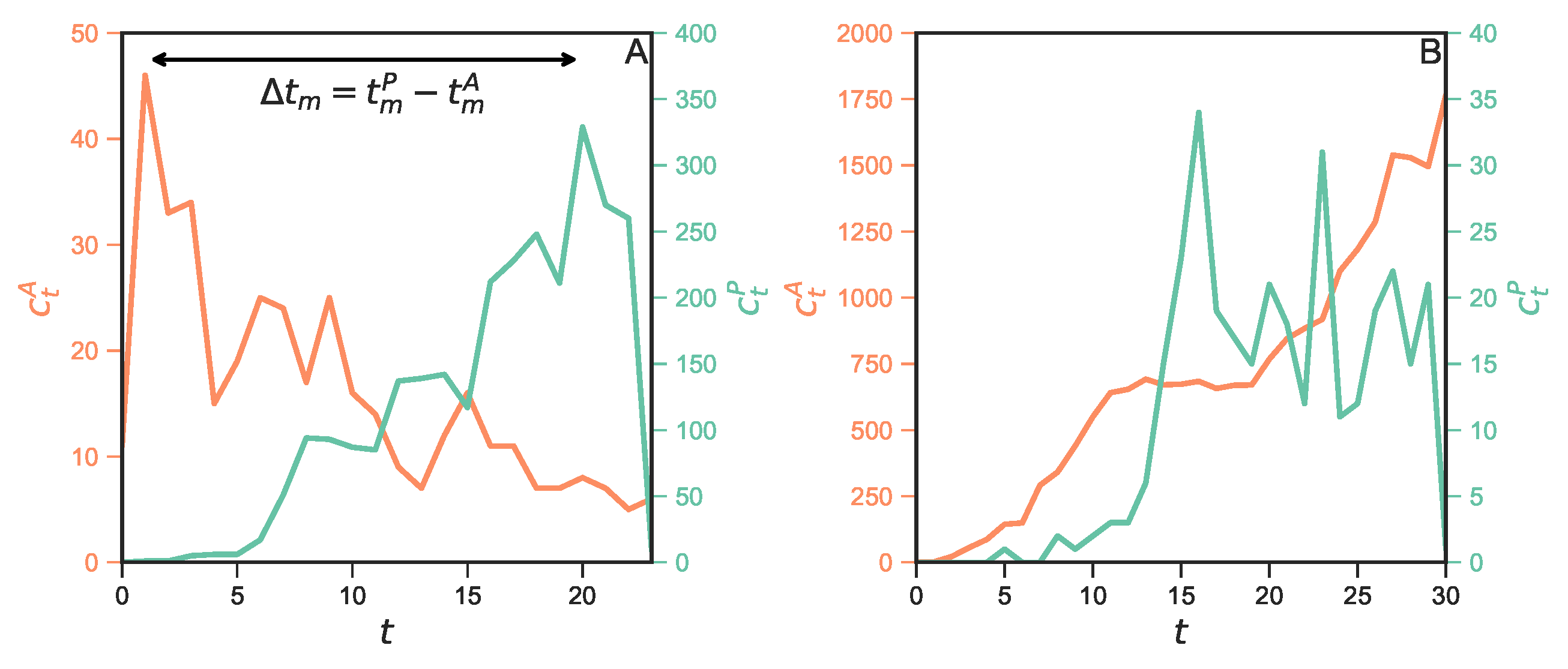 |
Comparing scientific and technological impact of biomedical research
Q Ke
Journal of Informetrics 12, 706-717 (2018)
arXiv |
abstract |
BibTeX
Traditionally, the number of citations that a scholarly paper receives from other papers is used as the proxy of its scientific impact. Yet citations can come from domains outside the scientific community, and one such example is through patented technologies—paper can be cited by patents, achieving technological impact. While the scientific impact of papers has been extensively studied, the technological aspect remains less known in the literature. Here we aim to fill this gap by presenting a comparative study on how 919 thousand biomedical papers are cited by U.S. patents and by other papers over time. We observe a positive correlation between citations from patents and from papers, but there is little overlap between the two domains in either the most cited papers, or papers with the most delayed recognition. We also find that the two types of citations exhibit distinct temporal variations, with patent citations lagging behind paper citations for a median of 6 years for the majority of papers. Our work contributes to the understanding of the technological impact of papers.
@article{ke2018comparing,
author = {Qing Ke},
title = {Comparing scientific and technological impact of biomedical research},
journal = {Journal of Informetrics},
volume = {12},
number = {3},
pages = {706--717},
year = {2018},
doi = {10.1016/j.joi.2018.06.010}
}
|
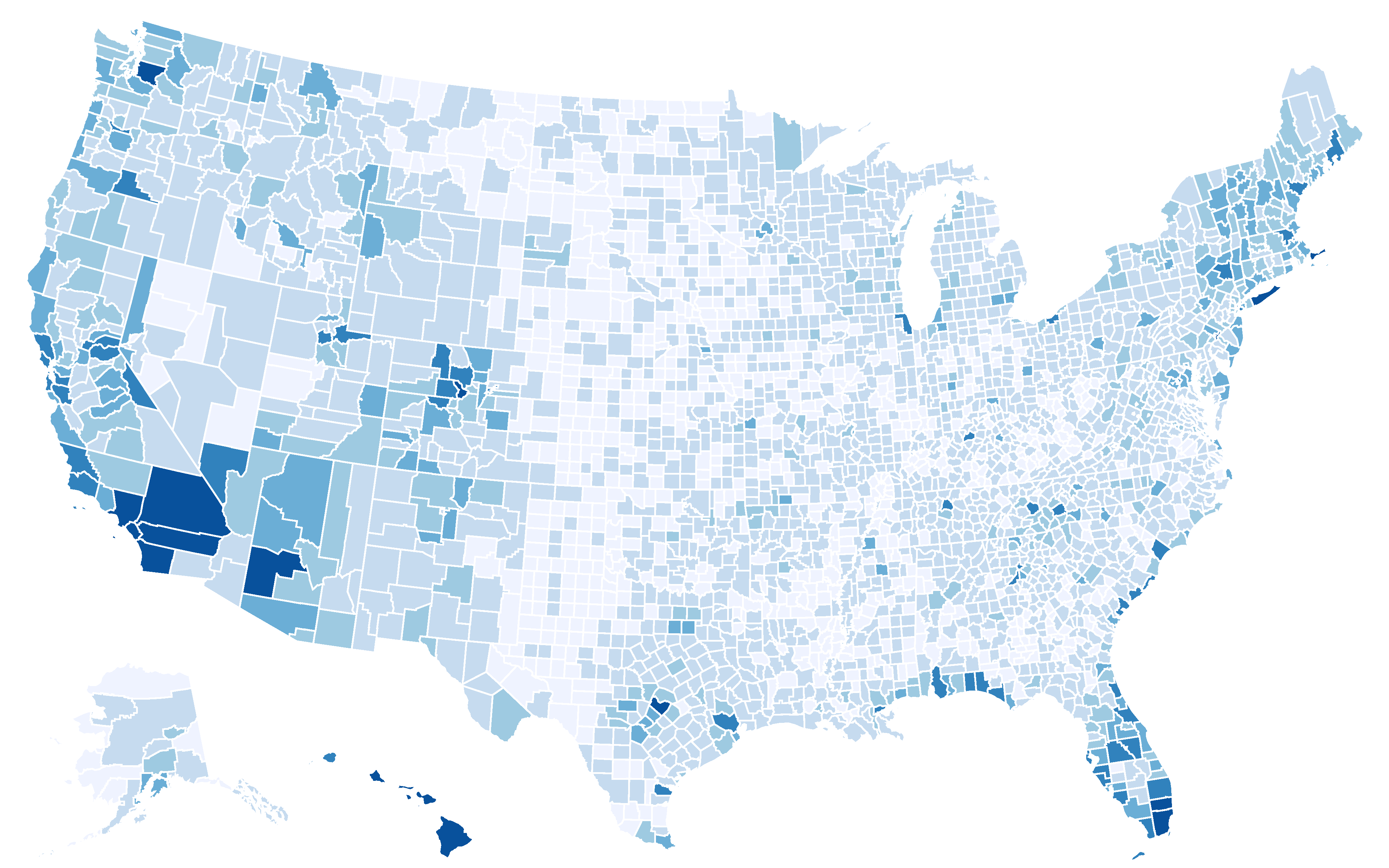 |
Service providers of the sharing economy: Who joins and who benefits?
Q Ke
Proceedings of the ACM on Human-Computer Interaction 1, 57 (2017)
arXiv |
abstract |
BibTeX |
Best Paper Honorable Mentions
Many "sharing economy" platforms, such as Uber and Airbnb, have become increasingly popular, providing consumers with more choices and suppliers a chance to make profit. They, however, have also brought about emerging issues regarding regulation, tax obligation, and impact on urban environment, and have generated heated debates from various interest groups. Empirical studies regarding these issues are limited, partly due to the unavailability of relevant data. Here we aim to understand service providers of the sharing economy, investigating who joins and who benefits, using the Airbnb market in the United States as a case study. We link more than 211 thousand Airbnb listings owned by 188 thousand hosts with demographic, socio-economic status (SES), housing, and tourism characteristics. We show that income and education are consistently the two most influential factors that are linked to the joining of Airbnb, regardless of the form of participation or year. Areas with lower median household income, or higher fraction of residents who have Bachelor's and higher degrees, tend to have more hosts. However, when considering the performance of listings, as measured by number of newly received reviews, we find that income has a positive effect for entire-home listings; listings located in areas with higher median household income tend to have more new reviews. Our findings demonstrate empirically that the disadvantage of SES-disadvantaged areas and the advantage of SES-advantaged areas may be present in the sharing economy.
@article{ke2017service,
author = {Qing Ke},
title = {Service Providers of the Sharing Economy: Who Joins and Who Benefits?},
journal = {Proc. ACM Hum.-Comput. Interact.},
volume = {1},
number = {CSCW},
year = {2017},
pages = {57:1--57:17},
doi = {10.1145/3134692}
}
|
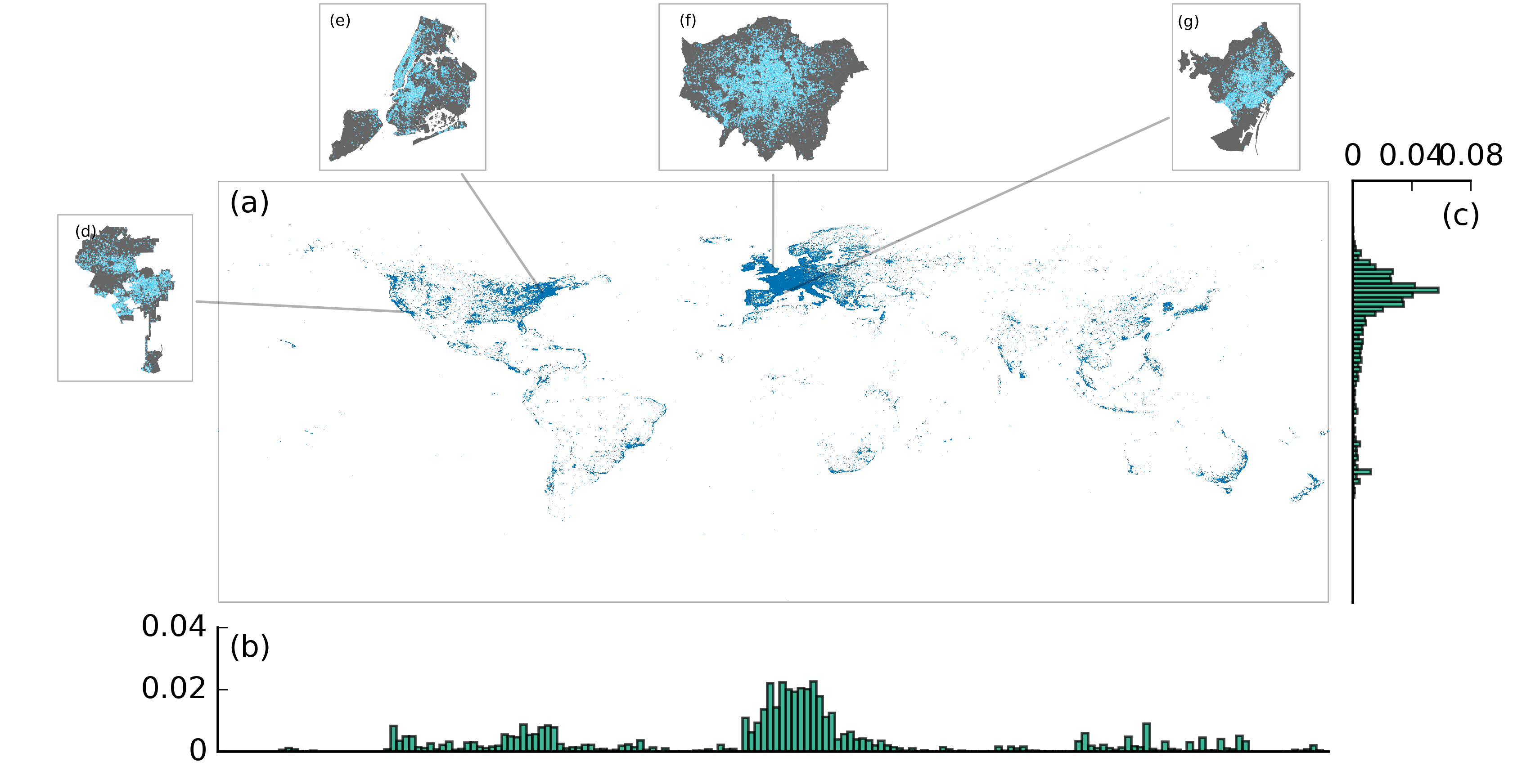 |
Sharing means renting?: An entire-marketplace analysis of Airbnb
Q Ke
In Proceedings of the 2017 ACM on Web Science Conference, 131-139.
arXiv |
abstract |
BibTeX
Airbnb, an online marketplace for accommodations, has experienced a staggering growth accompanied by intense debates and scattered regulations around the world. Current discourses, however, are largely focused on opinions rather than empirical evidences. Here, we aim to bridge this gap by presenting the first large-scale measurement study on Airbnb, using a crawled data set containing 2.3 million listings, 1.3 million hosts, and 19.3 million reviews. We measure several key characteristics at the heart of the ongoing debate and the sharing economy. Among others, we find that Airbnb has reached a global yet heterogeneous coverage. The majority of its listings across many countries are entire homes, suggesting that Airbnb is actually more like a rental marketplace rather than a spare-room sharing platform. Analysis on star-ratings reveals that there is a bias toward positive ratings, amplified by a bias toward using positive words in reviews. The extent of such bias is greater than Yelp reviews, which were already shown to exhibit a positive bias. We investigate a key issue—commercial hosts who own multiple listings on Airbnb—repeatedly discussed in the current debate. We find that their existence is prevalent, they are early-movers towards joining Airbnb, and their listings are disproportionately entire homes and located in the US. Our work advances the current understanding of how Airbnb is being used and may serve as an independent and empirical reference to inform the debate.
@inproceedings{ke2017sharing,
author = {Qing Ke},
title = {Sharing means renting?: An entire-marketplace analysis of Airbnb},
booktitle = {Proceedings of the 2017 ACM on Web Science Conference},
year = {2017},
pages = {131--139},
doi = {10.1145/3091478.3091504}
}
|
 |
A systematic identification and analysis of scientists on Twitter
Q Ke, YY Ahn, CR Sugimoto
PLoS ONE 12, e0175368 (2017)
arXiv |
abstract |
BibTeX |
Data
Nature News |
LSE Impact Blog
Metrics derived from Twitter and other social media—often referred to as altmetrics—are increasingly used to estimate the broader social impacts of scholarship. Such efforts, however, may produce highly misleading results, as the entities that participate in conversations about science on these platforms are largely unknown. For instance, if altmetric activities are generated mainly by scientists, does it really capture broader social impacts of science? Here we present a systematic approach to identifying and analyzing scientists on Twitter. Our method can identify scientists across many disciplines, without relying on external bibliographic data, and be easily adapted to identify other stakeholder groups in science. We investigate the demographics, sharing behaviors, and interconnectivity of the identified scientists. We find that Twitter has been employed by scholars across the disciplinary spectrum, with an over-representation of social and computer and information scientists; under-representation of mathematical, physical, and life scientists; and a better representation of women compared to scholarly publishing. Analysis of the sharing of URLs reveals a distinct imprint of scholarly sites, yet only a small fraction of shared URLs are science-related. We find an assortative mixing with respect to disciplines in the networks between scientists, suggesting the maintenance of disciplinary walls in social media. Our work contributes to the literature both methodologically and conceptually—we provide new methods for disambiguating and identifying particular actors on social media and describing the behaviors of scientists, thus providing foundational information for the construction and use of indicators on the basis of social media metrics.
@article{ke2017systematic,
author = {Qing Ke and Yong-Yeol Ahn and Cassidy R. Sugimoto},
title = {A systematic identification and analysis of scientists on Twitter},
journal = {PLOS ONE},
volume = {12},
number = {4},
pages = {e0175368},
year = {2017},
doi = {10.1371/journal.pone.0175368}
}
|
 |
Defining and identifying Sleeping Beauties in science
Q Ke, E Ferrara, F Radicchi, A Flammini
PNAS 112, 7426-7431 (2015)
arXiv |
abstract |
BibTeX |
Project
Nature Editorial |
Nature News |
NY Times |
Scientific American
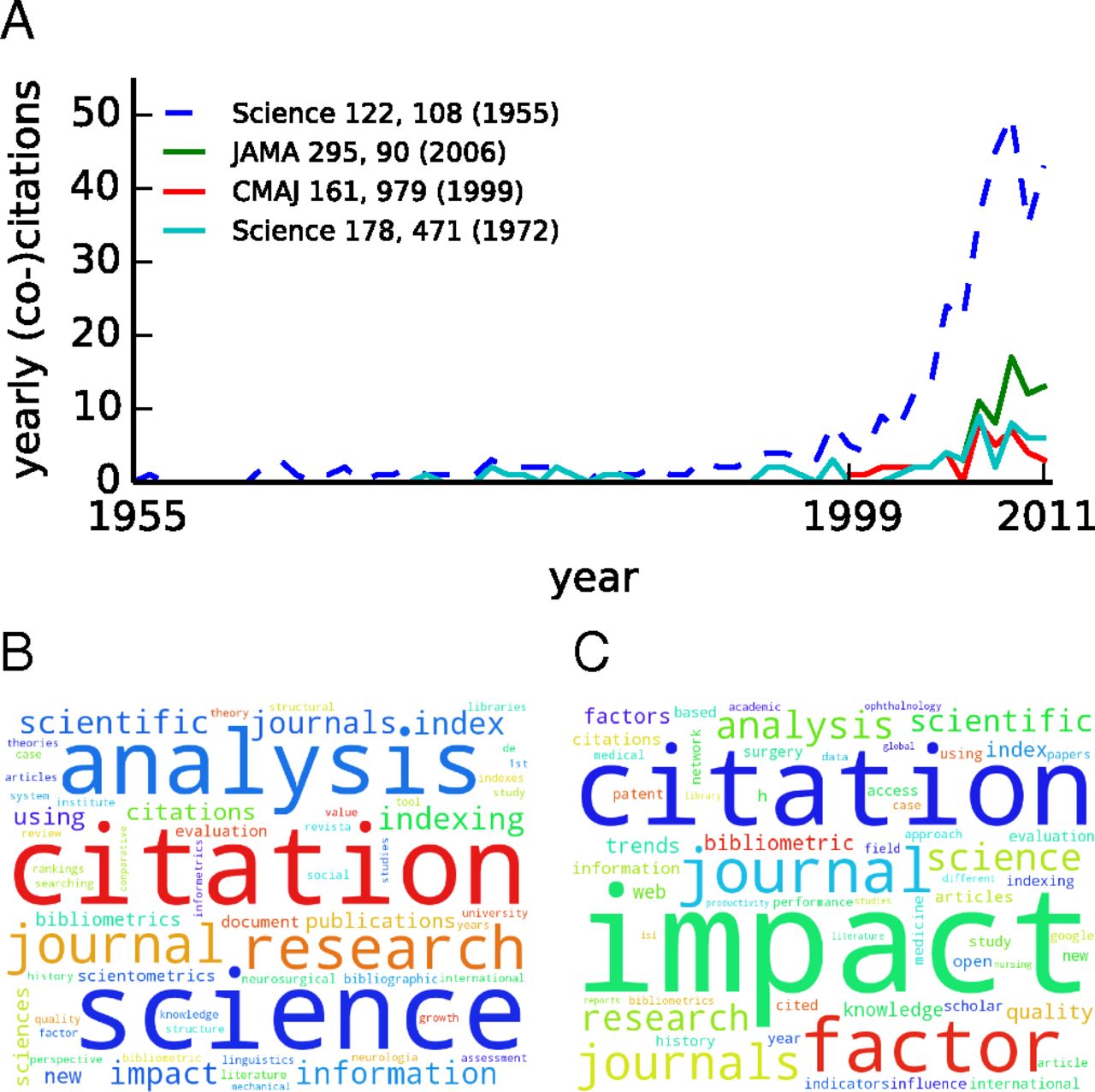
A Sleeping Beauty (SB) in science refers to a paper whose importance is not recognized for several years after publication. Its citation history exhibits a long hibernation period followed by a sudden spike of popularity. Previous studies suggest a relative scarcity of SBs. The reliability of this conclusion is, however, heavily dependent on identification methods based on arbitrary threshold parameters for sleeping time and number of citations, applied to small or monodisciplinary bibliographic datasets. Here we present a systematic, large-scale, and multidisciplinary analysis of the SB phenomenon in science. We introduce a parameter-free measure that quantifies the extent to which a specific paper can be considered an SB. We apply our method to 22 million scientific papers published in all disciplines of natural and social sciences over a time span longer than a century. Our results reveal that the SB phenomenon is not exceptional. There is a continuous spectrum of delayed recognition where both the hibernation period and the awakening intensity are taken into account. Although many cases of SBs can be identified by looking at monodisciplinary bibliographic data, the SB phenomenon becomes much more apparent with the analysis of multidisciplinary datasets, where we can observe many examples of papers achieving delayed yet exceptional importance in disciplines different from those where they were originally published. Our analysis emphasizes a complex feature of citation dynamics that so far has received little attention, and also provides empirical evidence against the use of short-term citation metrics in the quantification of scientific impact.
@article{ke2015defining,
author = {Qing Ke and Emilio Ferrara and Filippo Radicchi and Alessandro Flammini},
title = {Defining and identifying Sleeping Beauties in science},
journal = {Proceedings of the National Academy of Sciences},
volume = {112},
number = {24},
pages = {7426--7431},
year = {2015},
doi = {10.1073/pnas.1424329112}
}
|
 |
Tie strength distribution in scientific collaboration networks
Q Ke, YY Ahn
Physical Review E 90, 032804 (2014)
arXiv |
abstract |
BibTeX |
Data 1 |
Data 2 |
Code
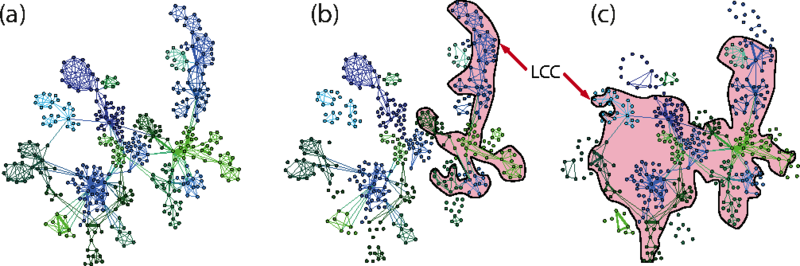
Science is increasingly dominated by teams. Understanding patterns of scientific collaboration and their impacts on the productivity and evolution of disciplines is crucial to understand scientific processes. Electronic bibliography offers a unique opportunity to map and investigate the nature of scientific collaboration. Recent work have demonstrated a counter-intuitive organizational pattern of scientific collaboration networks: densely interconnected local clusters consist of weak ties, whereas strong ties play the role of connecting different clusters. This pattern contrasts itself from many other types of networks where strong ties form communities while weak ties connect different communities. Although there are many models for collaboration networks, no model reproduces this pattern. In this paper, we present an evolution model of collaboration networks, which reproduces many properties of real-world collaboration networks, including the organization of tie strengths, skewed degree and weight distribution, high clustering and assortative mixing.
@article{ke2014tie,
author = {Qing Ke and Yong-Yeol Ahn},
title = {Tie strength distribution in scientific collaboration networks},
journal = {Physical Review E},
volume = {90},
issue = {3},
pages = {032804},
year = {2014},
doi = {10.1103/PhysRevE.90.032804}
}
|
















